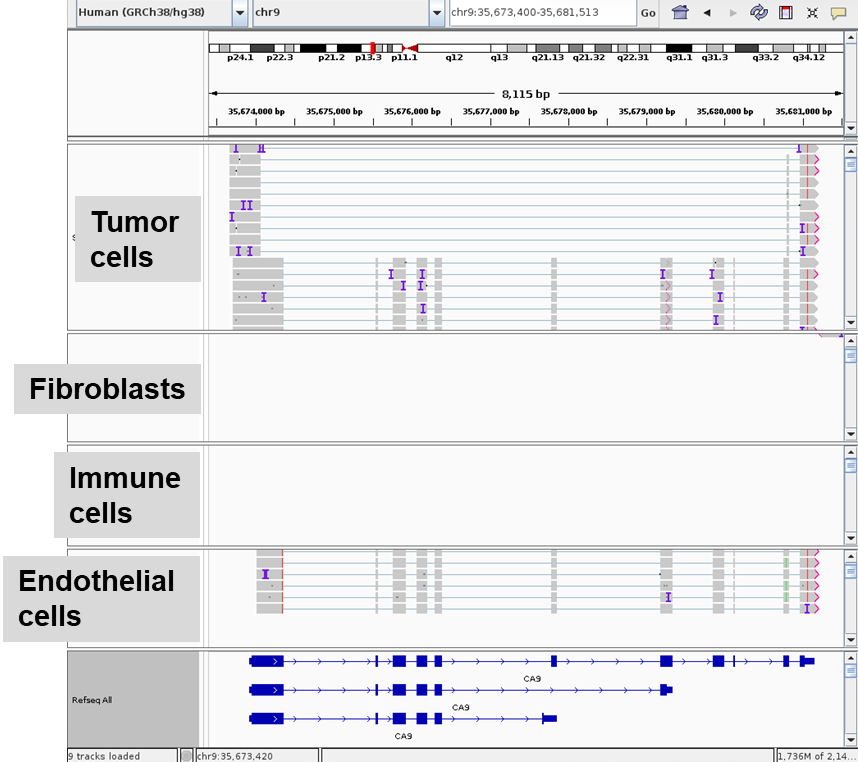
Alternative splicing is a mechanism by which a single gene can produce multiple transcripts and proteins with diverse functions. Following transcription, the pre-mRNA is processed by specialized mechanisms that cut out large segments of the transcript, leaving only exons (i.e., expressed regions) in the final mRNA transcript. From a functional point of view, a single cell may express multiple splice isoforms of the same gene, thereby conserving DNA storage resources.
Splicing is often related to cell phenotype - for example, mesenchymal and epithelial cells have distinct splice isoforms for many genes, allowing cells the flexibility and plasticity to transform from one cell type to another.
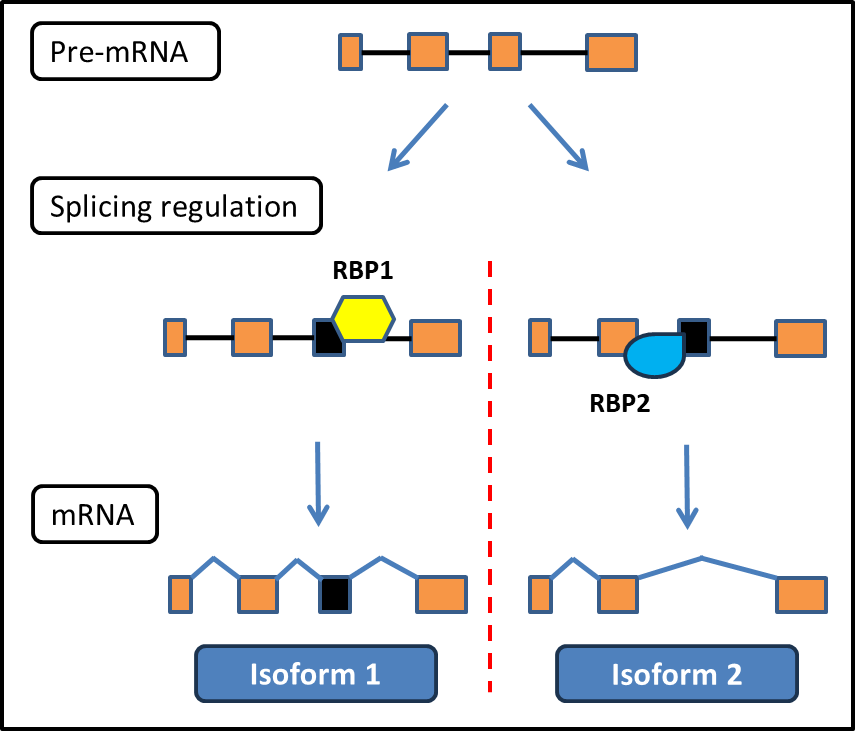
Splicing is carefully regulated during development and normal cell function. One mechanism of regulation involves RNA binding proteins (RBPs) that bind to pre-mRNA and promote exclusion or inclusion of specific exons in the final transcript.
Splicing is often disrupted in diseases such as cancer and neurodegenerative disorders, either due to mutations or to aberrations in regulatory mechanisms. New RNA-based therapeutic technologies are being developed to fix mis-spliced transcripts using antisense oligonucleotides, and will hopefully provide new ways to treat many diseases that are currently incurable.
In our lab, we aim to map the repertoire of spliced isoforms and uncover splicing regulation mechanisms in organ development, regeneration, and cancer. To this end, we use long-read single-cell RNA sequencing technologies to accurately capture full transcripts.