Biological systems must strike a delicate balance between robustness and adaptability. On the one hand, they need to resist noise and minor fluctuations to ensure stable development and function. On the other, they must remain responsive to changing environments, triggering appropriate biological programs such as regeneration or embryonic development. To meet both demands, cells rely on threshold-like logic: a system of internal switches that allows them to shift between distinct functional states in response to graded signals.
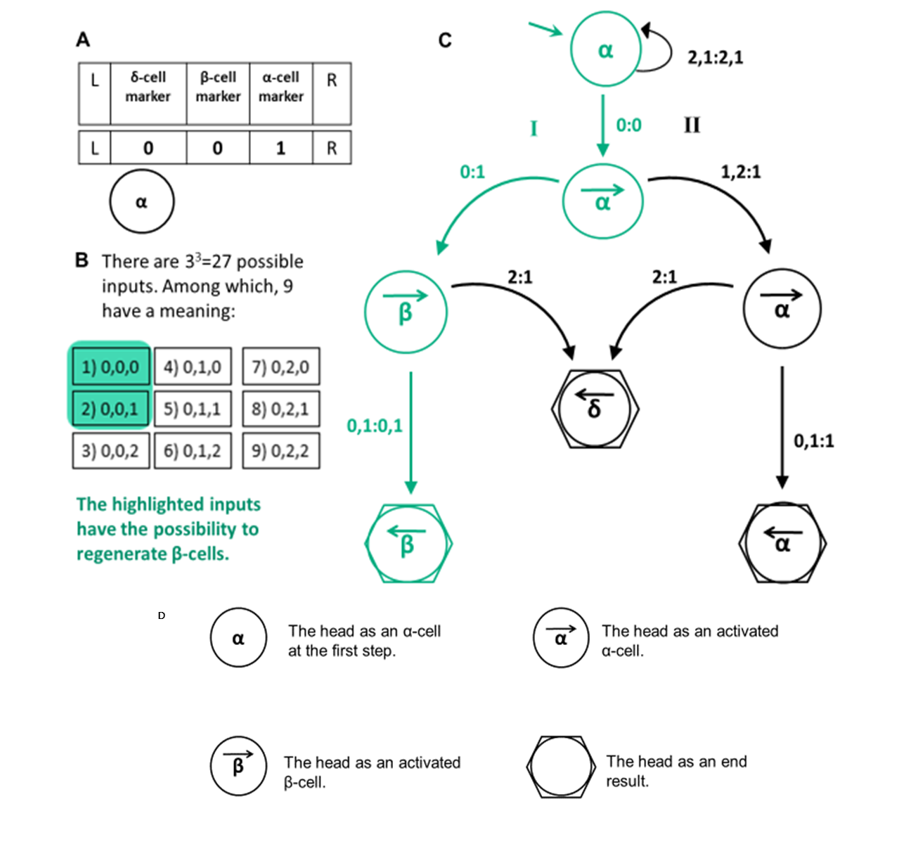
This concept is mirrored in experimental biology. When we manipulate genes, we typically lack fine control. We can reduce their expression through knockdown or knockout, overexpress them, or leave them untouched. These three experimental conditions (below, at, or above physiological levels) correspond naturally to a ternary system of values: 0, 1, and 2.
We believe that this three-valued logic is ideally suited to model biological processes. Our lab developed a mathematical framework based on this principle and uses it to analyze and predict complex biological responses. This approach has already led to important insights, including the prediction of a previously unknown signaling pathway involved in muscle formation and the identification of key conditions required for the emergence of insulin-producing pancreatic β-cells.
We continue to apply this framework to understand how cells make decisions in health, regeneration, and disease.